how to draw molecular orbital diagram
Molecular orbital diagrams provide qualitative information about the structure and stability of the electrons in a molecule. This article explains how to create molecular orbital diagrams in LaTdue eastTen by means of the package MOdiagram. For information about the more traditional molecular structure diagrams run into our documentation about chemical science formulae.
Introduction
Molecular diagrams are created using the surround MOdiagram. Below you lot can see the simplest working example:
\documentclass {article} \usepackage [utf8] {inputenc} \usepackage {modiagram} \begin {certificate} \begin {MOdiagram} \cantlet {left}{1s, 2s, 2p} \end {MOdiagram} \finish {document}
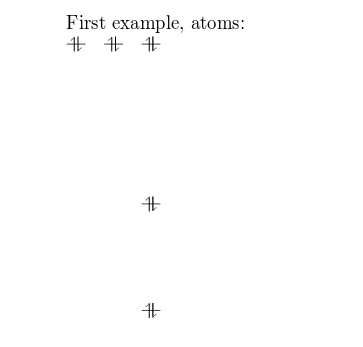
First, the package MOdiagrams is imported by
The basic command to draw MO diagrams is \atom. This command has two parameter in the example:
-
left. The alignment of the atom. -
1s, 2s, 2pAre the energy sub-levels to exist drawn. These can exist farther customized as you will larn in the next section.
Open an instance of the MOdiagram package in Overleaf
Atoms
You can pass some extra information nigh the atomic orbitals to the control presented in the introductory instance.
\brainstorm {MOdiagram} \atom {right}{ 1s = { 0; pair} , 2s = { 1; pair} , 2p = {1.v; up, down } } \atom {left}{ 1s = { 0; pair} , 2s = { 1; pair} , 2p = {one.5; upwardly, downwardly } } \end {MOdiagram}
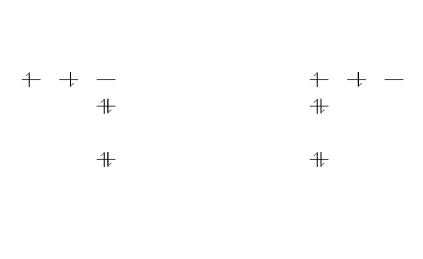
In this case, two identical atoms are drawn, left and correct-aligned respectively. The second argument in the command \atom contains some extra data. The generic syntax to create atoms is:
1s = {energy; specifications}
instead of 1s other possible values are 2s and 2p
- energy. Is the energy level, this number determines the vertical spacing in the diagram. If omitted is set to 0.
- specifications. A comma-separated list of the spins of the electrons contained in each orbital. The possible values are
up,down,pairand empty (only the semicolon is typed) for an empty orbital. If omitted is set topair.
For a better understanding of this syntax, beneath is a clarification of the commands in the previous example:
-
1s = { 0; pair}. The sub-level1sis in the0energy level, the orbital contains two (paired) electrons.
-
2s = { i; pair}. The sub-level2sfatigued in theanefree energy level, in that location are two electrons in this orbital.
-
2p = {1.5; up, down}. The sub-level2pdrawn in the free energy level1.5, i.east. in the diagram the vertical spacing is fix to 1.5; this sub-free energy level has two electrons: one with spinupwardlyin the first orbital and another with spindownwardsin the second orbital.
The same commands are repeated for the second atom on the correct.
Open up an case of the MOdiagram packet in Overleaf
Molecules
The syntax for molecules is very like to that of the \atom command presented in the previous department. The free energy sub-levels 1s, 2s and 2p become 1sMO, 2sMO and 2pMO respectively.
\begin {MOdiagram} \atom {left}{1s} \atom {right}{1s={;upward}} \molecule { 1sMO={0.75;pair,up} } \end {MOdiagram}
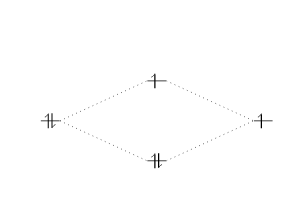
The syntax for \molecule takes two parameters just every bit the command \atom, but the orbital specifications are slightly unlike.
-
0.75is now the ratio (energy gain)/(energy loss). -
pair, upare the spins of the electrons in the bonding and anti-bonding molecular orbitals, respectively.
Below you tin can encounter a more elaborate instance to help understanding the syntax.
\brainstorm {MOdiagram} \atom {left}{ 1s, 2s, 2p = {;pair,up,upwards} } \atom {correct}{ 1s, 2s, 2p = {;pair,upwards,upward} } \molecule { 1sMO, 2sMO, 2pMO = {;pair,pair,pair,up,upward} } \end {MOdiagram}
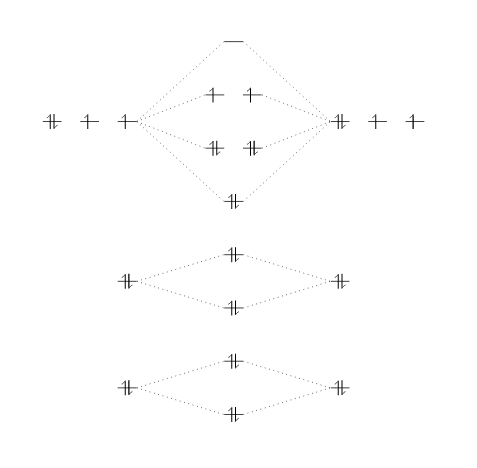
Three atoms are set on each side of the diagram and the respective molecule in the center.
Open an example of the MOdiagram bundle in Overleaf
Naming scheme
In that location is no straightforward way to set captions and names for elements in a MO diagram, but since this bundle is based on TikZ yous can use tikz commands within the MOdiagram environment:
\begin {MOdiagram} \atom {left}{1s} \atom {right}{1s={;up}} \molecule { 1sMO={;pair,up} } \draw [<-,shorten <=8pt,shorten >=15pt,blueish] (1sigma*) --++(ii,one) node {anti-bonding MO}; \end {MOdiagram}
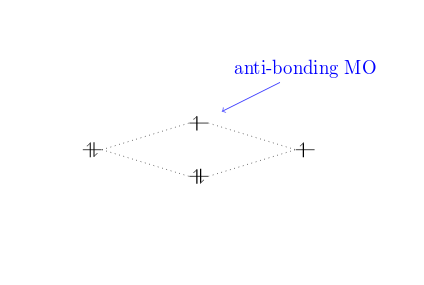
Notice that the proper name of the anti-bonding orbital 1sigma* can exist used for relative positions. Other labels bachelor are:
- 1sigma
- 2sigma
- 2pisigma
- 2piy
- 2piz
and the respective starred versions.
Open up an example of the MOdiagram packet in Overleaf
Farther reading
For more information run into:
- Chemistry formulae
- Feynman diagrams
- TikZ package
- Drawing Diagrams Directly in LaTeX
- Inserting Images
- List of Greek letters and math symbols
- The modiagram manual
- modiagram - a few examples
Source: https://www.overleaf.com/learn/latex/Molecular_orbital_diagrams
Posted by: bockmartyart49.blogspot.com


0 Response to "how to draw molecular orbital diagram"
Post a Comment